Where is Humanity Heading?
Naive opinions about the current status of human evolution follow two stereotypes - the "Big Brained Boffin" and "Evolution is dead! Long live evolution!" The actual picture is very different and shows evolution is ongoing apace, but with warnings for human culture, and its reproductive attitudes and technologies.
In the first scenario, it is assumed we will become ever more intellectualized, leading to individuals with huge brains and small bodies, possibly having to be born by cesarean section and likely selected by IVF as well, in a brave new world utopia. Some writers suggest that we might even become partly integrated with machines in a kind of cyborg fusion although this is biologically far-fetched science fiction.
The humpty-headed scenario is in obvious conflict with the fact that brain size has actually been getting a little smaller in the current epoch, despite our cultural explosion, after swelling from around 450 cc to 1500 cc with the emergence of early Homo species, with the reduction to a little over 1400 cc on average, possibly as a result of increases of overall efficiency of the brain's existing developmental plan and adaptive wiring.
In the second scenario, it is assumed that the advent of culture and medical science, with most people living to reproductive age, combined with contraception, has meant that any changes are a product of individual choices rather than natural selection and that cultural evolution has superseded natural evolution.
This is fallacious because it is clear that the capacity of prospective parents to determine the sex of their offspring, let alone avoid implanting embryos which might have genetic defects, or any characteristics the parents might find undesirable, or to seek embryos with genes that might be identified with anything from high intelligence, or good teeth to sexual orientation.
Moreover, it is clear from international demographics that contraception has led to extreme unevenness in reproductive rates with some populations, such as those of Taiwan, and Japan in impending rapid decline due to reproduction rates far below replacement, while in other countries such as Italy native European populations are failing to replace themselves due to career and lifestyle choices, while some immigrant populations are rapidly expanding due to much higher birth rates.
The role of religion and religious fundamentalism is also causing severe distortions to reproductive rates with Catholicism fighting a losing battle against contraception, while Muslim populations strive to populate through large families and in some cases multiple wives giving some Muslim men over 20 children and literally hundreds of grandchildren. To the extent that religious conservatism, or any other cultural trait, might also be partly based on the genetics of brain function, one can see that culture can both radically influence genetic selection and act as a feedback factor amplifying genes predisposed to cultural attitudes which promote reproduction in an era of contraception.
Since the Human Genome Project, our ability to get decisive data on human evolution has exploded, enabling us to not only trace our origins in gatherer-hunter populations of Africa and following the radiative diversification of human peoples, but also to discover key evolutionary changes, both confluent with the emergence of culture and accompanying human migration to new habitats (Sabeti et. al. 2006 Positive Natural Selection in the Human Lineage 312 1614-20).
In a study of genetic markers in 270 people from four groups Han Chinese, Japanese, Yoruba and Northern Europeans Henry Harpending and John Hawks found at least 7% of human genes had undergone evolution in the last 5,000 years, including the ability of adults to digest milk, the LARGE gene conferring resistance to Lassa fever in Yoruba , partial resistance to other diseases such as malaria, changing skin pigmentation, including changes that assist survival in northern latitudes through improving vitamin D generation in the skin. They estimate the contemporary evolutionary rate to be as much as 100 times faster than at any time since the split of the earliest hominin from the great apes (Ward, P 2009 What will become of Homo sapiens? Sci. Am. Feb).
Indeed many genetically derived medical conditions in current human populations appear to be adaptive responses to epidemic disease or environmental stress. High cholesterol, like light skin enhances vitamin D production, since vitamin D is made from light splitting steroids. Hemochromatosis, the accumulation of too much body iron, is related to white blood cells, which happen also to regulate iron intake, that may have adapted to increase resistance to plague bacteria which enter and attack the immune system through the iron metabolism of these white blood cells. By losing its iron regulation, the immune cells protected against the entry of the plague but also through their lack of iron ended up signaling to the rest of the body not to excrete any iron. This scenario has been made somewhat notorious in a popular book (Sharon Moalem Survival of the Sickest: A Medical Maverick Discovers Why We Need Disease). It was paradoxically confirmed when Dr. Malcolm Casabadan died while researching a strain of yersinia pestis weakened by disabling its iron metabolism. The argument goes that, when introduced into his body, through inadequate laboratory protection, the weakened iron-deficient strain feasted on his disproportionately iron-rich bloodstream.
Sickle cell anemia, a recessive condition that appears in African and African American people carrying two copies of the defective gene, likewise confers resistance to malaria in individuals carrying a single copy of the allele. A notable example of a selective sweep is the gene variant discovered which gives Tibetans living at high altitudes better adjustment of red-cell production, which spread through the population in the last 3000 years (Xin Yi 2010 Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude Science 329 75-80).
Despite the advent of medical science, it is likely that disease resistance will continue to shape human evolution. Unless and until an effective vaccine is found for HIV there will be selection pressures favouring the survival of more resistant individuals and emergence of resistant genotypes. The continuing risk of multiply antibiotic resistant epidemic diseases from TB to gonorrhea could likewise lead to the emergence of new genotypes. If the world population remains as long-lived as currently in developed countries, it is likely that we may also see increased resistance to cancer emerge. Although senescence is not usually accompanied by reproductive procreation cancer is becoming an increasing mortality factor in all age groups as other diseases become controlled.
Many of the genetic changes which have taken place during human diversification were due to changing habitats with migration, that in addition, subjected populations to new diseases, and to major changes with urbanization, with new assaults due to poor sanitation, often inadequate diets and epidemic diseases, and reproductive changes that may have accompanied the emergence of culture, including language proficiency and social impacts on reproductive rates caused by cultural forces.
Modern society is tending to homogenize much of the diversification which occurred as human groups spread out across the Earth, forming new cultures and adapting in different ways to new environments, so is it undoing the evolutionary process that accompanied our adaptive radiation?
A more extreme version of the "evolution is dead" scenario asserts that contraception is promoting evolution in reverse, because people unwilling or unable to cope with contraception are more likely to conceive. People with genetic damage that was once fatal, or prevented conception, are now able to live and have children. More endowed individuals, who seek higher education and careers, tend to put off having families, while less endowed people, who pay less heed to contraception, or long term family planning, reproduce early and prolifically. However there is little real evidence that overall intelligence is decreasing in societies where it is measured.
Nevertheless, some conditions which we do find socially disadvantageous could easily become more common in the current social evolutionary climate. It has been controversially suggested by David Comings, that Tourette's syndrome and ADHD have become more common because women with these conditions are less likely to attend college and more likely to reproduce.
On the other hand, some people have gone so far as to claim that intelligence is not heritable because it is a product of a large number of small genetic differences and cannot easily be selected for, but this argument is completely fallacious, as social intelligence is the trademark of our species and a dominant portion of measured human evolution consists of small changes in a large number of interacting genes having a cumulative effect, rather than rapid selective sweeps of one, or a few genes, which alone confer a marked increase in the viability or number of offspring.
A selective sweep results when a single gene, or a small collection of closely linked genes which would travel together as 'hitchhikers' with meiotic crossing over of nearby chromosomal regions have a significant selective advantage in the next generation. This can result in a gene which start out at say 1% frequency growing over a small number of generations to say 99% frequency because all the offspring which possess it have say a 10%increase in their offspring, causing an exponential rise. Such a selective sweep would take about 200 generations or around 5000 years. For example the lactase gene that allows adults to digest milk rather than switching off when a child is weaned spread in the dairy farming populations of Europe in 5000 to 10000 years. A parallel rapid evolution in lactase has independently occurred in African dairy farming populations.
However the evolutionary evidence coming out of recent genetic analyses shows that, although there are a few outstanding examples of selective sweeps, most of the evolutionary changes are multifactorial, involving a large number of genes conferring only a small selective advantage, taking a much longer time for such evolutionary change. Many of the changes discovered happened earlier in the human spread out of Africa in response to local environments, such as the genes favoring lighter skin, and were then carried to other places only slowly being changed by relatively neutral evolution as changing circumstances altered selection pressures (Pritchard J. 2010 How we are evolving Sci. Am. Oct).
In a study of 1000 individuals, from around the world, investigating single nucleotide polymorphisms (SNPs), which indicate selective pressure when random variation in a population is suppressed into one or a few dominant types by natural selection, selective sweeps appear to date from three distinct events. The first is the immediate aftermath of the radiation out of Africa, when human populations were still confined to the Middle East around 60000 years ago. There are two further sweeps, a West Asian series occurring at high frequency in populations of Europe, Middle East, Central and South Asia, but not elsewhere, and a further series confined to East Asian, Native Americans, Melanesians and Papuans both occurring around 30000 to 20000 years ago.
Moreover, genes which earlier conferred selective advantage and became common in selective sweeps have not reappeared when the same conditions occur in other migrating populations. For example the SLC24A5 gene conferring lighter skin colour in people of Northern Europe is not found in significant quantities in East Asians where in Northern climates other genes promote skin lightness.
The genes which show greatest contrasts between populations indicating selective adaption do not show the hitchhiking signals of neighbouring regions common in selective sweeps, but rather the gradual propagation over tens of thousands of years associated with multi-factorial gene effects involving tens to thousands of interacting genes which confer only a small selective advantage on any singe gene.
This is characteristic of many significant features of human evolution from height to intelligence, yet the fact that pygmy populations of African rain forest adapted to become small demonstrates that selection changing the frequency of individual genes by only a few percent but changing many genes simultaneously can have a significant adaptive effect.
At the end of this section are three sections looking in detail at some of the key findings concerning human evolution and genetic changes as a result of the human and other genome projects in relation to our (1) human differentiation from the apes, (2) our differentiation from and occasional interbreeding with sister species such as Neanderthals and (3) the detailed divergence of modern human groups in the migrations out of Africa. These give a more in depth insight into the diversity of genetic evidence that has emerged in these key areas of discovery and include several examples of genes hypothetically linked to human intelligence and the emergence of culture.
So what of the future? In modern society where medical science can compensate for many of the effects of disease and environment on life expectancy and fertility, one would expect selective advantage to be operating most acutely on factors which effect human reproduction, or on genes which affect social attitudes to reproduction, including those affecting social intelligence.
Stephen Stearn and his team have identified six different heritable traits in women that are associated with higher lifetime numbers of children, including being slightly shorter and stouter and having a late menopause. One of the central genetic explanations for male homosexuality, giving a 14% genetic basis on its own, is the fertile mother effect - that a gene which confers increased fertility in women also carries an increased likelihood of male offspring becoming homosexual, but not enough to offset the increased fertility (Stearn S et. al. 2010 Measuring Selection in Contemporary Human Populations. Nature Reviews Genetics 11 611-622).
Heritable characteristics affecting attitudes to reproduction could also be pivotal in determining human evolutionary futures. Religiosity has been claimed to have a 40% genetic component, which can be explained readily as an adaptive response enabling the formation of larger more dominant societies through reducing intra-social defection from the group. Major religions, especially the Judeo-Christian-Islamic nexus all have a primary aim of encouraging unconstrained reproduction to further the utopian aims of the religion. There is room here for substantial concern that cultural and genetic factors are combining to have an undue strategic influence on the human genetic and evolutionary future.
A second social factor which is highly significant to human evolution is sexual rather than natural selection. Geoffrey Miller has drawn attention to the beneficial effects of sexual selection and mutual mate choice in driving the emergence of art, music, science, culture and the evolution of super-intelligence in Homo sapiens, through sexual selection, particularly by astute females selecting partners with good husbanding, bringing home the bacon, or an antelope in good hunting, telling stories and jokes around the camp fire and adroit musicianship, as genuine indicators of genetic fitness (Miller G The Mating Mind).
In our work (Fielder C and King C Sexual Paradox: Reproductive Conflict and Human Emergence http://sexualparadox.org) we extend this idea to look at the effects of patriarchal dominance on sexual selection on a variety of contexts, from hypergamy and the dowry, through to sexual imbalances caused by invocations against female reproductive choice and selective female infanticide. The dominance of patriarchy has also led to significant changes in adaptive advantage, for example favouring higher testosterone women in high-born places who give birth to more male children, and the opposite in low born populations where marrying up a daughter to a high born son carries more advantage.
 Venus of Laussel attests to the importance of fertility, understanding of the menstrual period (13 notches on the moon horn) and an emphasis on the feminine as sacred in early human populations.
Venus of Laussel attests to the importance of fertility, understanding of the menstrual period (13 notches on the moon horn) and an emphasis on the feminine as sacred in early human populations.Patriarchy may also be inhibiting some of the central features of mammalian evolution by inhibiting female reproductive choice, often by dire punishments such as stoning for adultery, which is usually selectively aimed at female indiscretion and loss of honour. Only three percent of mammals are socially monogamous, largely due to the fact that they bear live young thus causing the females to have a massive investment in parenting, while the males tend to invest primarily or exclusively in sexual fertilization. In many ways Homo sapiens is at at extreme with a massive pregnancy straining the females resources and carrying significant risks of mortality so female reproductive choice should be at a premium in our species.
This is compounded with X-Y sexual determination, which means that, particularly in terms of intelligence bearing genes, males express a unique X in their brain while females express a tortoise-shell chimera of paternal and maternal X. Thus female selection of well-endowed males serves as a principal selective factor for X-linked genes associated with brain function, of which there are several, confirmed by the greater variation in natural intelligence in human males than females.
The relentless discovery of genes which not only cause medical conditions, but also affect our behavior is exposing us all to the desire to avoid disadvantageous genes in our offspring and select offspring through IVF and pre-implantation genetic diagnosis (PGD) that have favored chosen characteristics. Millions of baby girls are already aborted as a result of ultrasound scans in countries where people favor boys over girls, leading to serious sex imbalances.
While we have tended to leave human selection to individual choice humanity has for centuries been involved in directed evolution of our food plants and domestic animals, so it is only a matter of time before we enter into an era of selective breeding and germ-line engineering of the human species as well. In a sense religious prerogatives to 'go forth and multiply' have been a form of socially induced selective breeding that has become more pronounced since the advent of contraception.
All of these prospects bring with them major pitfalls. Many species we have applied selective breeding to have lost critical viability factors such as disease resistance and sexual viability. Cauliflowers have lost natural resistance to mosaic virus and Cavendish bananas have become almost monoclonal. Bulldogs have become dependent on Caesarian section birth and world salmon stocks are becoming contaminated by genetically-modified salmon farming varieties (GM Salmon Muscle In on Wild Fish When Food Is Scarce Sci. Am. 8 Jun 2004).
Humans are entering a phase where cesareans, often for cosmetic reasons, are rising upwards towards 40% of live births, compromising the future capacity of humans to give live natural birth. IVF combined with intracytoplasmic sperm injection (ICSI) is resulting in transmission of sperm defects which would render a man infertile from fathers to their sons. We need to take heed from the mistakes we have made with other species and not commit humanity to threats to our viability as a result of the over-technologization of human reproductive capacity in a Brave New World utopia.
At the same time, genetic testing is placing new pressures on human viability in terms of withholding of medical insurance on the basis of a propensity to chronic or terminal disease. There are also implicit dangers in society entering into genetic control of human reproduction. Societies which have embraced eugenics have almost without exception resulted in atrocities and massive human injustices.
Central to the future of humanity, as an intelligent species, is ensuring the capacity of women, who are responsible for bringing the next generation into existence, to choose their partners, free of religious or paternal pressures, and giving them encouragement and the cultural values to make these choices carefully and astutely, by choosing male partners to sire their children who can demonstrate the mental and physical prowess which accompanies a good functioning set of genes, as well as socially conscious attitudes and resourceful parenting.
Finally we should respect human diversity and avoid aiming for any single Brave New World utopia and resist religious and social prerogatives that attempt to flood the world with believers, or cause massive imbalances in reproduction rates, and are in their essence, both selfish and distorting.
We are in an era where the human population is cresting to unprecedented levels and the resulting impacts on the Earth's capacity to sustain us. Long term human viability is being threatened by climate change, non-renewable resource depletion, species and genetic loss and habitat destruction. Far from a technological utopia, the pressures of economic decline in an era of post peak oil price spirals, humanity could find itself again facing a struggle of tooth and claw amid pressures of world domination by fundamentalist reactions to poverty and hardship and a necessity to cope without the luxuries of advanced medical care as many people struggling in developing countries still have to do today.
We thus need to limit human population to sustain future human viability and share this duty of care between religious believers and rationalists and the well-endowed and otherwise. The lesson of the Neanderthals is that genetic diversity coming from individuals we might think are 'inferior' may be essential to the viability of our own offspring.
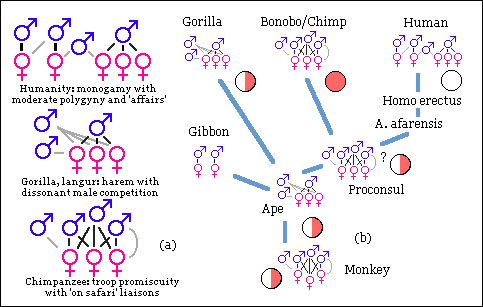 Primate mating patterns and concealed or advertised ovulation.
Primate mating patterns and concealed or advertised ovulation.Pink and white discs indicate overt and covert ovulation, half-shaded slight signs.
Evolutionary Features of Human Emergence that Differ from Apes
Genes involving disease resistance, reproduction (seminal protamines and seminogelins), and nocioception (awareness of pain and toxic substances) stand out as rapidly evolving. Gene evolution is faster on rearranged chromosomes 1, 2, 5, 9, 12, 15, 16, 17, 18. Rapidly evolving gene clusters are associated with immunity, host defense, chemosensation and inflammation. Hominid lines show increased divergence of genes associated with ion transport, neruotransmission, sound perception and reproduction by comparison with murids. Large gene families such as those involved in immunity and olfactory are harder to test but are also subject to accelerated divergence.
Six regions of low diversity have been noted, consistent with linked genes hitchhiking on strong selective sweeps in recent human history. In addition one region containing several high diversity-divergence scores contains genes noted for selective mutations, FoxP2 involved in speech ( Suite of chatterbox genes discovered New Scientist 11 Nov 2009, Nature, DOI: 10.1038/nature08549), as well as CFTR (connected with asthma resistance). Other selective sweeps have also been discovered in recent human evolution putatively associated with brain size determining genes microcephalin and ASPM (see below).
Looking at regulators and genes which have been lost in the human lineage one prominent one removes a forebrain subventricular zone enhancer near the tumour suppressor gene growth arrest and DNA-damage inducible, gamma (GADD45G)11,12, a loss correlated with expansion of specific brain regions in humans. (McLean CY et. al. 2011 Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471/7337, 216-9).
Many brain genes are more strongly conserved than other genes, although brain evolution may be affected by new alternative splicing arrangements in a small subset of genes, and strong selective sweeps made by new highly selective genes. Gene conversion with a pseudogene for example in the human line has altered sialic acid expression in brain tissue so that only human microglia express sialic acid.
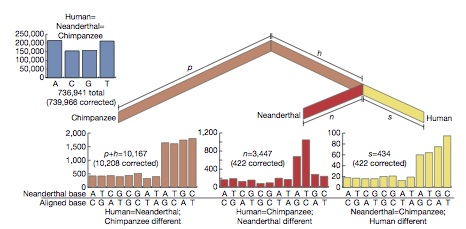
A detailed map of human evolution is emerging from comparison between the results of the human and chimp genome projects (Chimpanzee sequencing and analysis consortium Initial sequence of the chimpanzee genome and comparison with the human genome Nature 437 69-87). The discoveries so far are summarized below:
The single nucleotide divergence between chimp and human is only 1.23% (35 million base pairs) of which 1.06% corresponds to fixed fixed divergence between the species once 14-20% of intra-species polymorphisms are taken into account. The correspondingly higher divergence of the Y of 1.9% and lower of the X of 0.94% indicates the male mutation rate is 3-6 times the female rate. This mutational discrepancy between the sexes supports the necessity of female reproductive choice in all hominins, including humans.
Regarding mutation generally, CpG (cytosine-guanine) bases are prone to deamination to TpG and constitute the dominant form of chemical damage, causing 25.2% of mutations but only occurring in 2.1% of bases. Mutation rate variations between sexes are not CpG correlated because chemical damage is time related as opposed to replication errors from the 5-6 fold higher number of cell divisions in spermatogenesis. Mutation rates also vary with location with 15% more near the ends of chromosomes, with higher local recombination rate, high gene density and high GC content. This effects the shorter chromosomes more significantly. Dark bands which are gene poor have a 10% higher mutation rate. Telomere regions have a higher rate rate in hominids than in murids, suggesting less selective pressure.
Insertions and deletions are rarer individually than point mutations but are larger, so 1.5% of the euchromatic differences are species specific, a larger difference than point mutations. Overall indel divergence is about ~45 Mb in each species or about 3% of the genome dwarfing the single nucleotide divergence. Most are very small, but the total contribution is 73% from larger over 80bp indels. Over a third of these come from repetitious micro-satellite and satellite sequences, a quarter are caused by transposable elements, with a residue coming from deletions in the other species. 8.3 Mb of these in humans contain 34 exon regions indicative of new genes.
Orthologous proteins are very similar between the species, with 29% identical. Recombination is limited by a low cross-over rate with 1kb only having only ~1 crossover every 100,000 generations, or 2 million years.
Estimates of the effects of natural selection, neutral evolution and other factors can be gleaned by examining KA the amino acid substituting mutations, KS the synonymous mutations and KI the non-coding mutations. KA is 37% higher in the most distal 10 Mb than in proximal regions so careful averaging across the genome is needed. Both KA/KS and KA/KI are around 0.23 <<1 indicating significant purifying selection. This value is 35% greater than for murids (rodents) indicating either greater positive adaption or fewer evolutionary constraints. There is also weak purifying selection on silent sites in exons compared with introns In terms of gene evolution a KA/KS of 0.23 indicates 77% of amino acid substitutions are sufficiently deleterious to be eliminated by natural selection. 4.5% of human-chimp orthologues (585/13,454) have KA/KL<1 indicating strong selection, however because of the low divergence between the species, about half could occur by chance variation among genes. The high KA/KS for these genes also shows that 25% of amino acid substitutions contribute to the current human genetic load. The KA/KS for human polymorphisms of 0.20-0.23 is very similar to that for human chimp divergence, indicating little positive selection across the genome driving evolutionary divergence, because positive selection would cause many selective sweeps and reduce polymorphisms, indicating fewer evolutionary constraints in the hominid lines than in murids. X has KA/KL = 0.32 skewed high and low with higher selection on testis-related genes. The median is similar to autosomes indicating a skewed subset with very high evolutionary selection. Many low values indicate greater purifying selection consistent with being genes expressed in the hemizygous single X state in males, and high values could also result from positive selection from adaptive hemizygosity of the X in males, particularly if a substantial proportion of these genes are recessive.
Transposable elements are now known to play a significant part in evolutionary change. The history of transposable elements differs significantly between the species. Endogenous retroviruses have died out in humans except for HERV-K with 73 human insertions (of which 66 have only the long terminal repeat remaining, indicating old insertions) This occurs also in chimps 45 insertions (44 LTR only) but chimps have other active ERVs. PtERV1 has over 200 copies over half of which are full length indicating active insertion. SINEs in particular Alu has been 3 times more active in humans (7000/2300 lineage specific insertions) mostly due to two new subfamilies (Ya5 Ya8), but baboons are 1.6 times higher still. Old Alu elements lie in GC rich gene-rich regions but newer ones are in AT rich gene-poor regions where LINEs also accumulate, consistent with L1-based retro-transcription. There is similar L1 activity in both species (~2000 insertions), as well as 200 human and 300 chimp processed LINE-related retrogenes (pseudogenes). SVA an Alu repeat with a CpG island and potential transcription factor binding sites occurs (~1000) in both species. 3 human genes show SVA insertions resulting in species differential transcriptions. There are also 612 human and 914 chimp Alu-Alu deletions, 26 and 48 L1 deletions and 8, and 22 LTR deletions, none involving exons of human genes in chimp. There are also larger scale inversions and fusions. Chimp chromosomes 12,13 now called 2A,2B fused to make human chromosome 2. There have also been pericentric inversions.
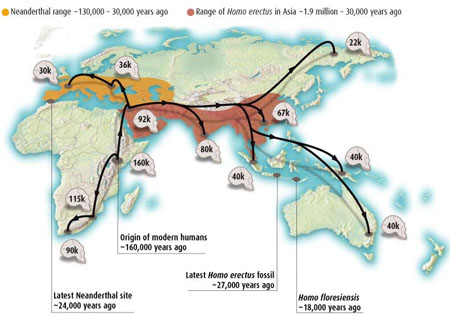
 According to initial genetic analysis, Neanderthals diverged from homo sapiens ~500,000 years ago. There has been no major interbreeding, but possibly some transfer of genes e.g. from human males to Neanaderthal females, although candidate human genes conferring natural advantage do have a profile consistent with transfer from Neanderthals (Green R,et. al. 2006 Analysis of one million base pairs of Neanderthal DNA Nature 444 330-336. doi:10.1038/nature05336, Science 314, 1068-71).
According to initial genetic analysis, Neanderthals diverged from homo sapiens ~500,000 years ago. There has been no major interbreeding, but possibly some transfer of genes e.g. from human males to Neanaderthal females, although candidate human genes conferring natural advantage do have a profile consistent with transfer from Neanderthals (Green R,et. al. 2006 Analysis of one million base pairs of Neanderthal DNA Nature 444 330-336. doi:10.1038/nature05336, Science 314, 1068-71).Divergence from and Interbreeding with Neanderthals and Denisovians
More recent comprehensive investigation of the Neanderthal genome (Green, R. E. et al. 2010 Science 328, 710-722., Nature | doi:10.1038/news.2010.225) suggests that there was a period of interbreeding between Neanderthals and humans in the Near East around the time of the first migration out of Africa, rather than more recently in Europe, as the putative sequences are shared by non-African French, Han and Papuan, but not by the African Yoruba or San. It is estimated that among the former, 1-4% of the genome derives from Neanderthal sequences, although there is little evidence for these corresponding to the specific genes suggested by Lahn's team. Other transfers could have occurred but are not apparent in the research.
The "Out of Africa" hypothesis may be consistent with a degree of regional development involving some sexual interbreeding with Neanderthals and Homo erectus. Specific genes such a s PDHA1 consist of two families with the last common ancestor 1.8 million years ago, and microcephalin variants appearing 40,000 years ago (p 105) also have differences suggesting an original divergence 1 million years ago suggesting 'introgression' from Neanderthals (Jones, Dan 2007 The Neanderthal Within New Scientist 3 Mar.). An even more ancient divergence in the pseudogene RRM2P4 in East Asian people suggests interbreeding with Homo Erectus. Some evidence from skeletons is also consistent with this picture. However more recent sequencing of the Neanderthal nuclear genome (Callaway E 2010 Neanderthal genome already giving up its secrets New Scientist 06 May) suggests little or no interbreeding with Homo sapiens and has cast doubt on the existence of the microcephalin variant in Neanderthals, as well as a gene associated with increased fertility in Icelanders also attributed to transfer from Neanderthals.
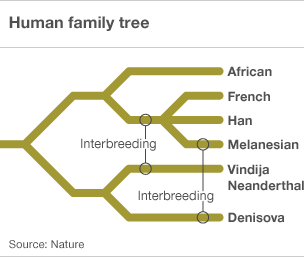
However, research in 2010 into microsatellite variation in the human genome suggests that there have been two interbreeding events with other hominid species, one about 60,000 years ago in the eastern Mediterranean after the emergence of Homo sapiens from Africa, and the other about 45,000 years ago in eastern Asia (Nature | doi:10.1038/news.2010.194, Krause, J. et al. Nature doi:10.1038/nature08976 ).
More recently we have the discovery of remains of Denisovians (Callaway E 2010 Fossil genome reveals ancestral link Nature 468, 1012 doi:10.1038/4681012a, Finlayson C 2010 All change: Theories of human ancestry get an overhaul BBC 31 Dec ) a further species branching off from the Neanderthals. Genetic analysis of the remains indicates a significant interbreeding specifically with Melanesian people of some 6% (Reich D, et. al. 2010 Genetic history of an archaic hominin group from Denisova Cave in Siberia Nature doi:10.1038/nature09710).
More recent investigations show that interbreeding with other hominins was critical to the globalization of Homo sapiens. Human leukocyte antigens (HLAs), a family of about 200 genes that essential to our immune system also contains some of the most variable human genes: hundreds of versions - or alleles - exist of each gene in the population, allowing our bodies to react to a huge number of disease-causing agents and adapt to new ones. One allele, HLA-C*0702, is common in modern Europeans and Asians but never seen in Africans; Peter Parham has found it in the Neanderthal genome, suggesting it made its way into H. sapiens of non-African descent through interbreeding. HLA-A*11 had a similar story: it is mostly found in Asians and never in Africans, and Parham found it in the Denisovan genome, again suggesting its source was interbreeding outside of Africa. This tallies with interbreeding giving Homo sapiens pivotal resistance to non-African diseases. While only 6 per cent of the non-African modern human genome comes from other hominins, the share of HLAs acquired during interbreeding is much higher. Half of European HLA-A alleles come from other hominins, says Parham, and that figure rises to 72 per cent for people in China, and over 90 per cent for those in Papua New Guinea (Marshall M. 2011 Breeding with Neanderthals helped humans go global New Scientist 16 Jun, Neanderthal sex boosted immunity in modern humans BBC 26 Aug 2011).
There is also suggestive evidence for up to 2% interbreeding with another hominin species, in ancient African populations of Biaka Pygmies and San Bushmen (Human ancestors interbred with related species Nature | doi:10.1038/news.2011.518, Hammer, M. F. et al. Genetic evidence for archaic admixture in Africa PNAS http://dx.doi.org/10.1073/pnas.1109300108) although this is based only on statistical divergences in some loci, and lacks a sister species reference sequence, it suggests interbreeding around 35,000 years ago with a population that originally diverged from the Homo sapiens line some 700,000 years before.
Genetic Diversification of Modern Humans
Chromosomes contain a variety of markers that can be used to compare diverse populations and infer an evolutionary relationship between them. These include the slowly varying protein polymorphisms of coding regions which are useful for long-term trends, single nucleotide polymorphisms, and non-coding region changes (mutation rates about 2.5 x 10-8 per base pair per generation and useful for reconstructing evolutionary history only over millions of years) insertion and deletion events (about 8% of polymorphisms, extending from one to millions of nucleotides), particularly those driven by transposable elements such as the LINEs and even more frequent SINEs such as Alu, non-coding micro-satellites (mutation rate 10-5 - 10-2 due to repeat slippage) and mini-satellite regions of repeating DNA (mutation rates as high as 2 x 10-1 due to meiotic recombination in sperm) that both evolve rapidly and are not subject to the strong selection of coding regions which can differentiate changes over the much shorter time scales of modern human migration (See: Sexual Paradox The Entanglements of Biological Sex http://sexualparadox.org/biology.htm).
The insertions and deletions of the million or so Alu elements in the human genome are particularly useful, as the most active sub-population of about 1000 Alu is actively transcribing and undergoing rapid change. A subpopulation of Alu are capable of generating new coding regions (exons), when inserted into non-coding introns between spliced sections of a translated mRNA, because one base-pair change within Alu leads to formation of a new exon reading into the surrounding DNA. This is not necessarily deleterious because alternative splicing still allows the original protein to be made as well. We have the highest number of introns per gene of any organism, and thus have to have gained an advantage from this costly error-prone process. Alus may have given rise, through alternative splicing, to new proteins that drove primates' divergence from other mammals. Recent studies have shown that the nearly identical genes of humans and chimps produce essentially the same proteins in most tissues, except in parts of the brain, where certain human genes are more active and others generate significantly different proteins through alternative splicing of gene transcripts. Our divergence from other primates may thus be due in part to alternative splicing.
Highlighting unique features of human genetic evolution, are two key genes whose mutations cause microcephaly, consistent with increased brain size, whose rapid spread through the human population may coincide with spurts in human culture. Microcephalin appeared ~37,000 years ago (Evans,P; et. al. Microcephalin, a Gene Regulating Brain Size, Continues to Evolve Adaptively in Humans Science 309 1717-20) coinciding with the birth of culture and ASPM spread from the Near East around 5000 years ago (Mekel-Bobrov, et. al. Ongoing Adaptive Evolution of ASPM, a Brain Size Determinant in Homo sapiens Science 309 1720-2). However studies linking these variants have failed to find differences in intelligence and results remain highly controversial (DOI:10.1126/science.314.5807.1872). Nevertheless, these results are consistent with an overall examination of linkage disequilibrium in single nucleotide polymorphisms (Moyzis R et. al. 2005 PNAS doi: 10.1073/pnas.0509691102) which indicate that about 7% of our genes have been subject to selection in the last 50,000 years, a figure similar to domestication of maize, including genes for protein metabolism, disease resistance and brain function.
The persistence of polygyny is also manifest in the greater divergence between human groups in the X-chromosome than other chromosomes, caused by women possessing double X and men only a single. The diversity arises because some men don't get to pass on their genes, while most women do. When Michael Hammer and his colleagues sequenced DNA from 90 people belonging to six groups: Melanesians, Basques, Han Chinese, as well as three African cultures: Mandenka, Biaka and San. Hammer's team discovered more genetic differences in the X chromosome than would be expected if equal numbers of males and females tended to mate, over human history. The only explanation for this pattern is widespread, long-lasting polygyny (PLoS Genetics DOI: 10.1371/journal.pgen.100202).
If we consider the likely effects of the out of Africa hypothesis, we would expect that founding African populations not subject to active expansion and migration would have greater genetic diversity and that the genetic makeup of other world populations would come from a subset of the African diversity, consisting of those subgroups who migrated.
In the case of mitochondrial mtDNA (mutation rate about 2.5 x 10-7) and its hyper-variable D-loop (mutations rates as high as 4 x 10-3), which is transmitted only down the maternal line (see Tishkoff Sarah, Verrelli Brian 2003 Patterns of Human Genetic Diversity: Implications for Human Evolutionary History and Disease Annu. Rev. Genomics Hum. Genet. 4:293–340 for caveat) and the non-recombining majority of the Y-chromosome which is transmitted only down the paternal line, each with no recombination, we would expect greater diversity going deeper into the historical tree of divergence, with certain existing groups who have retained the founding patterns of survival and have not undergone rapid population expansions to retain an increasingly diverse source variation. All these features are broadly observed in the genetic data to date.
 (a) MtDNA tree for African groups showing haplotypes of !Kung, Mbuti and Biaka as well as the line coming out of Africa (Chen Y, et. al. 2000 mtDNA Variation in the South African Kung and Khwe - and Their Genetic Relationships to Other African Populations Am. J. Hum. Genet. 66:1362–1383). (b) Diagram of world migration and regional differentiation of successive mtDNA haplotypes (Gilbert Tom 2003 Death and Destruction New Scientist 31 May 32). (c) mtDNA distances between founding African groups including Hadza (clicks) Khwe is from (Knight A, et. al. 2003 African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages Current Biology, 13, 464–473). Recent mtDNA evidence suggests a first wave of migration down the coast of Asia all the way to Australia (Forster P., Matsumura S 2005 Did early humans go north or south? Science 308 965-6).
(a) MtDNA tree for African groups showing haplotypes of !Kung, Mbuti and Biaka as well as the line coming out of Africa (Chen Y, et. al. 2000 mtDNA Variation in the South African Kung and Khwe - and Their Genetic Relationships to Other African Populations Am. J. Hum. Genet. 66:1362–1383). (b) Diagram of world migration and regional differentiation of successive mtDNA haplotypes (Gilbert Tom 2003 Death and Destruction New Scientist 31 May 32). (c) mtDNA distances between founding African groups including Hadza (clicks) Khwe is from (Knight A, et. al. 2003 African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages Current Biology, 13, 464–473). Recent mtDNA evidence suggests a first wave of migration down the coast of Asia all the way to Australia (Forster P., Matsumura S 2005 Did early humans go north or south? Science 308 965-6).Most studies of non-coding regions of autosomal, X-chromosome, and mitochondrial mtDNA genetic variation (which are desirable markers because they are not so subject to selection and thus have relatively neutral drift) show higher levels of genetic variation in African populations compared to non-African populations, using many types of markers. Although some studies of Y-chromosome variation have observed higher heterozygosity levels in non-African populations, the African populations have higher levels of pairwise sequence differences, consistent with these populations being ancestral. High levels of diversity in African populations alone do not prove that African populations are ancestral. A recent bottleneck event and/or colonization and extinction events among non-African populations, or a more recent onset of population growth in non-Africans, could also cause a decrease in genetic diversity (Tishkoff and Verrelli). In fact the complete inter-fertility of all human populations and the relative lack of genetic divergence by comparison with the few remaining chimp colonies in the wild (Hrdy, Sarah Blaffer 1999 Mother Nature : A History of Mothers, Infants, and Natural Selection Pantheon New York 183) does indicate a significant bottleneck. The genetic data is consistent with a human emergence from a population of only 10,000 around 100,000 years ago. This is also consistent with the delayed maturation, long birth spacings as a result of prolonged lactation and high infant mortality seen in gather-hunter populations such as the !Kung. At such low growth rates a population of 100 would take 50,000 years to reach 10,000 (Hrdy 183).
 Patterns of male migration. The Genographic Project - a partnership between National Geographic and IBM - will collect DNA samples from over 100,000 people worldwide to provide a high-resolution genetic map of human migration.
Patterns of male migration. The Genographic Project - a partnership between National Geographic and IBM - will collect DNA samples from over 100,000 people worldwide to provide a high-resolution genetic map of human migration.However studies of protein polymorphisms as well as mtDNA haplotypes, X-chromosome and Y-chromosome haplotypes, autosomal microsatellites and minisatellites, Alu elements, and autosomal haplotypes indicate that the roots of the population trees constructed from these data are composed of African populations and/or that Africans have the most divergent lineages, as expected under a recent African origin rather than a multi-regional emergence model. Additionally, studies of autosomal, X-chromosomal haplotype and mtDNA variation indicate that Africans have the largest number of population-specific alleles and that non-African populations harbor a subset of the genetic diversity that is present in Africa, as expected if there was a genetic bottleneck when modern humans migrated out of Africa. Analysis of genetic variation among ethnically diverse human populations indicates that populations cluster by geographic region (i.e., Africa, Europe/Middle East, Asia, Oceania, New World) and that African populations are highly divergent. The mtDNA studies hypothesize a primal female ancestor - the African Eve - around 150,000 years ago (Chen et. al. see earlier) while the Y-chromosome Adam is more recent, at around 90,000 years ago (Underhill P, et. al. 2001 The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations Ann. Hum. Genet., 65, 43-62) consistent with the greater reproductive variance of males than females. Differences between the Y- and mtDNA distributions indicate how migration, intermarriage and female exogamy have affected the gene pool. The genetic patterns of both these and autosomal microsatellites (Zhivotovsky, L., Rosenberg N, Feldman M 2003 Features of Evolution and Expansion of Modern Humans, Inferred from Genomewide Microsatellite Markers Am. J. Human Genetics May) are consistent with founding African diversity with migratory radiations to form other world populations, with deep founding radiations to the forest people such as the Biaka and Mbuti, Khoisan click-language speaking !Kung-san bushmen of Botswana and the Sandawe of Tanzania, and possibly the Hadzabe, as well as the forest people such as the Mbuti and Biaka 'pygmies' who have adopted the Bantu languages of the farming neighbours with which they now share semi-symbiotic relationships. Along with some Ethiopian and Sudanese sub-populations, these groups may represent some of the oldest and deeply diversified branches of modern humans.
 Genographic project study of mitochondrial origins shows a deep split separating Khoisan mitochondrial inheritance from other groups, including those migrating out of Africa, and a deep division between two Khoisan types L0k and L0d going back 140,000 years, suggesting a separation of some 100,000 years possibly caused by long term drought in Africa (Behar et al., 2008 The Dawn of Human Matrilineal Diversity, The American Journal of Human Genetics , doi:10.1016/j.ajhg.2008.04.002).
Genographic project study of mitochondrial origins shows a deep split separating Khoisan mitochondrial inheritance from other groups, including those migrating out of Africa, and a deep division between two Khoisan types L0k and L0d going back 140,000 years, suggesting a separation of some 100,000 years possibly caused by long term drought in Africa (Behar et al., 2008 The Dawn of Human Matrilineal Diversity, The American Journal of Human Genetics , doi:10.1016/j.ajhg.2008.04.002).Such recent genetic evidence has laid bare the relationships between some of the founding human groups spread across Africa from the 'Cushite' horn of Ethiopia to the southern Kalahari. Mitochondrial DNA studies have highlighted the ancient origin of the !Kung San and of pygmy peoples of the Congo Basin such as the Mbuti and the Biaka.
Y-chromosome studies have shown the !Kung share a most ancient haplotype with sub-populations from Ethiopia and the Sudan. According to an overall survey of genetic research by Sarah Tishkoff of the University of Maryland, the most deeply ancestral known human DNA lineages may be those of East Africans, such as the Sandawe, who share many phenotypic features and a click language with the !Kung. This suggests southern Khoisan-speaking peoples originated in East Africa. The most ancient populations are now believed to also include the Sandawe, Burunge, Gorowaa and Datog people of Tanzania. The Burunge and Gorowaa migrated to Tanzania from Ethiopia within the last 5,000 years consistent with an ancient founding population in this area. Echoes of the earliest language spoken by ancient humans tens of thousands of years ago may have been preserved in the distinctive clicking sounds still spoken by some existing African tribes.
The persistence of polygyny is also manifest in the greater divergence between human groups in the X-chromosome than other chromosomes, caused by women possessing double X and men only a single. The diversity arises because some men don't get to pass on their genes, while most women do. When Michael Hammer and his colleagues sequenced DNA from 90 people belonging to six groups: Melanesians, Basques, Han Chinese, as well as three African cultures: Mandenka, Biaka and San. Hammer's team discovered more genetic differences in the X chromosome than would be expected if equal numbers of males and females tended to mate, over human history. The only explanation for this pattern is widespread, long-lasting polygyny (PLoS Genetics DOI: 10.1371/journal.pgen.100202).
Highlighting unique features of human genetic evolution, are two key genes whose mutations cause microcephaly, consistent with increased brain size, whose rapid spread through the human population may coincide with spurts in human culture. Microcephalin (Evans-Gilbert et. al.) appeared ~37,000 years ago coinciding with the birth of culture and ASPM spread from the Near East around 5000 years ago (Mekel-Bobrov et. al.) However studies linking these variants have failed to find differences in intelligence and results remain highly controversial (DOI:10.1126/science.314.5807.1872). Nevertheless, these results are consistent with an overall examination of linkage disequilibrium in single nucleotide polymorphisms (Moyzis et. al.) which indicate that about 7% of our genes have been subject to selection in the last 50,000 years, a figure similar to domestication of maize, including genes for protein metabolism, disease resistance and brain function.
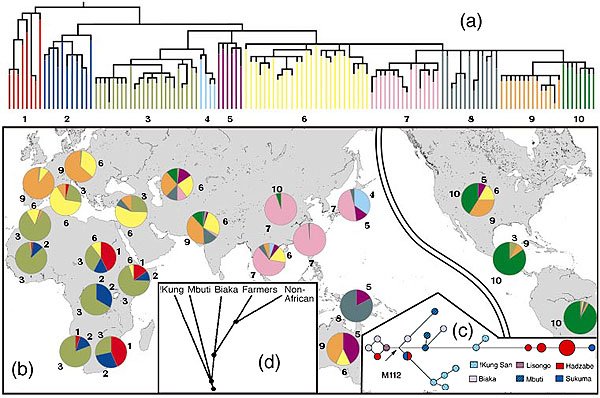 (a) Non-recombining Y-chromosome evolutionary tree (Underhill P, et. al. 2001 The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations Ann. Hum. Genet., 65, 43-62) (b) Geographical distribution showing the ancient haplotype shared by the San and Ethiopian and Sudanese sub-populations. (c) Genetic distances between Khoisan and forest peoples sharing M112 a Y-chromosome allele common only in these groups showing great genetic distance between Hadzabe and San peoples (Knight A, et. al. 2003 African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages Current Biology, 13, 464–473) . (d) Autosome satellite analysis confirming ancient divergence of San and forest peoples leading to migration from Africa (Zhivotovsky, L., Rosenberg N, Feldman M 2003 Features of Evolution and Expansion of Modern Humans, Inferred from Genomewide Microsatellite Markers Am. J. Human Genetics May).
(a) Non-recombining Y-chromosome evolutionary tree (Underhill P, et. al. 2001 The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations Ann. Hum. Genet., 65, 43-62) (b) Geographical distribution showing the ancient haplotype shared by the San and Ethiopian and Sudanese sub-populations. (c) Genetic distances between Khoisan and forest peoples sharing M112 a Y-chromosome allele common only in these groups showing great genetic distance between Hadzabe and San peoples (Knight A, et. al. 2003 African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages Current Biology, 13, 464–473) . (d) Autosome satellite analysis confirming ancient divergence of San and forest peoples leading to migration from Africa (Zhivotovsky, L., Rosenberg N, Feldman M 2003 Features of Evolution and Expansion of Modern Humans, Inferred from Genomewide Microsatellite Markers Am. J. Human Genetics May).
In a counterpoint to these studies, Rohde and coworkers (Rohde D. L. T, Olson S. & Chang J. T. 2004 Modelling the recent common ancestry of all living humans Nature, 431, 562- 565, Hein J 2004 Pedegrees for all humanity Nature 431, 518-9) estimate that the repeated spreading of family trees by sexually recombining mobile populations and differences in reproductive rates leads to an estimate of the most recent common ancestor of our global populations existing just 3,500 years ago, excepting these most isolated groups.
 Human divergence trees calculated by single nucleotide polymorphisms (SNPs) top left (Jun Z. Li,et al. 2008 Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation Science 319 1100 DOI: 10.1126/science.1153717) bottom right Jakobsson et. al. (Mattias Jakobsson et. al. 2008 Genotype, haplotype and copy-number variation in worldwide human populations Nature 451 998 doi:10.1038/nature06742). Trees for haplotypes and copy number variation between populatons (Jun et. al.).
Human divergence trees calculated by single nucleotide polymorphisms (SNPs) top left (Jun Z. Li,et al. 2008 Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation Science 319 1100 DOI: 10.1126/science.1153717) bottom right Jakobsson et. al. (Mattias Jakobsson et. al. 2008 Genotype, haplotype and copy-number variation in worldwide human populations Nature 451 998 doi:10.1038/nature06742). Trees for haplotypes and copy number variation between populatons (Jun et. al.).